ANUBI: A Platform for Affinity Optimization of Proteins and Peptides in Drug Design
Imagine we could tinker with the building blocks of life, crafting proteins and peptides with the knack to bind more tightly to disease targets, like finding the perfect key for every lock. The quest for better drugs is much like searching the cosmos: vast, intricate, and, until now, slow. But the researchers behind ANUBI have built a new kind of tool—a digital compass guiding us across the landscape of possibility, mapping paths where better medicines might lie.
TL;DR
- ANUBI automates the search for protein and peptide drugs with improved binding, predicting closer matches in weeks, not months.
- The platform merges smart simulation and sequence design, saving time and cost over standard experimental drug screens.
Drug design often begins with a coveted question: how well does a candidate molecule stick to its biological target? In the world of proteins and peptides—molecules made of chains of amino acids—this is a puzzle with countless possible answers. Traditional methods test one possibility after another in the lab, but this process is slow and expensive, like fishing in a vast ocean with a single line.
Computational models have started to help. Programs like AutoDock Vina and learning-based tools can screen thousands of small molecules quickly, but when it comes to larger, more flexible protein-to-protein or protein-to-peptide interactions, they struggle. These connections aren’t just about static snapshots; they are more like intricate dances, where tiny changes can yield big differences in binding strength. Most existing scoring methods don’t catch these subtleties.
ANUBI (Anubi Nexus for Understanding Binding Interactions) tries to remedy this gap. Its creators describe a two-part computational platform: one part explores the vast sequence space—different possible protein or peptide variants—using a Monte Carlo engine (a method that, borrowing from the world of gambling, makes smart random changes and keeps the winners). The other part calculates how tightly each new candidate would grasp its target, using advanced molecular dynamics (MD) simulations combined with a method called MMPBSA to estimate binding free energies. This dual strategy allows ANUBI to mimic, at digital speed, the selection processes that might take months or years in the real world, and it does so with minimal input and expertise required from users beyond providing an initial molecular model.
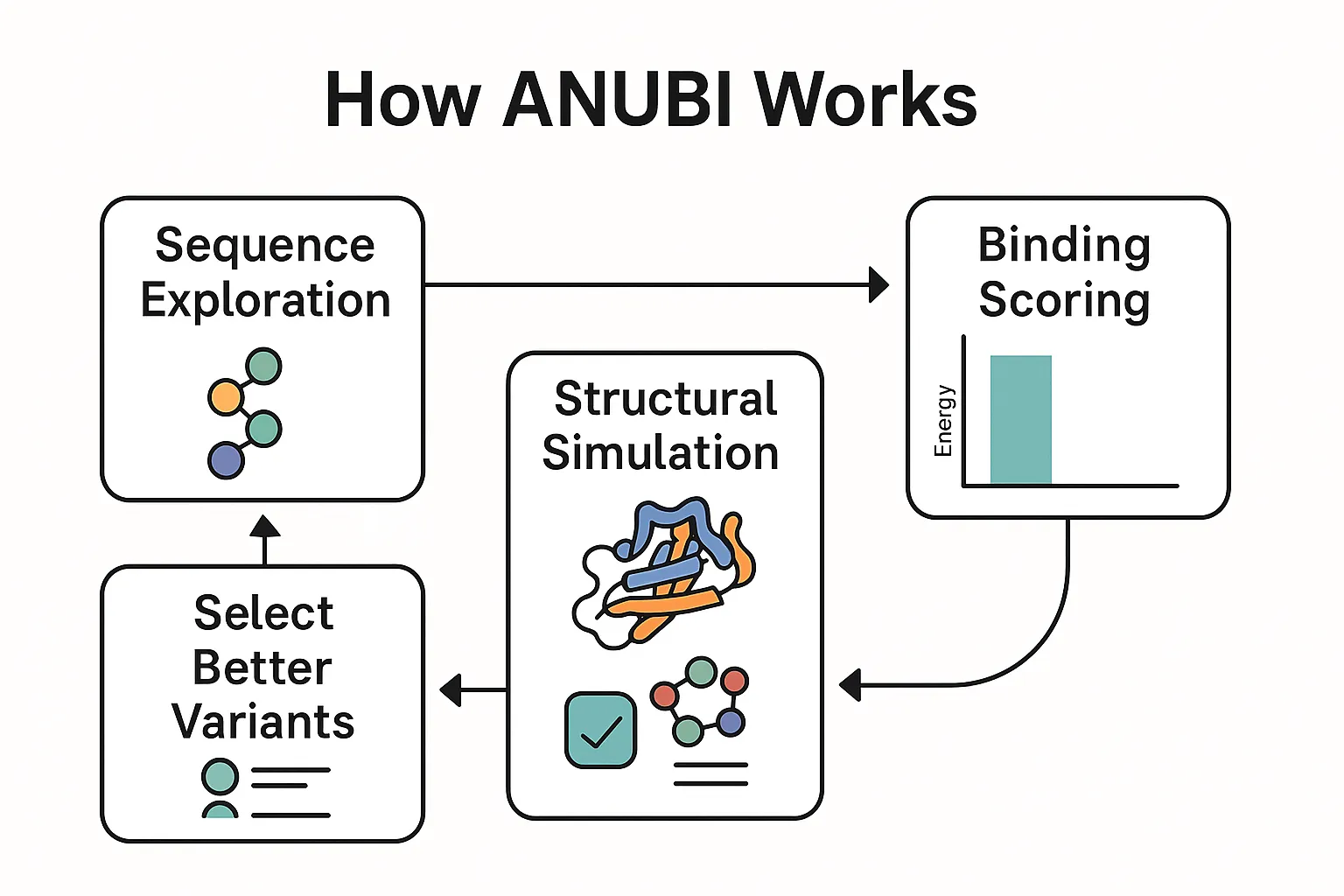
Figure 1. Schematic overview of the ANUBI platform.
(1) The sequence exploration engine proposes new protein/peptide variants.
(2) Molecular dynamics simulations examine each candidate’s 3D behavior.
(3) MMPBSA-based binding scoring estimates how tightly the molecule binds its target.
(4) High-scoring variants are selected and fed back into the next round of exploration.
When put to the test, ANUBI was set loose on two sample cases: an antibody-antigen interaction and a peptide-protein complex—both classic targets in modern drug discovery. Within about twenty days of computational exploration running on a standard GPU, ANUBI generated dozens of candidate molecules, identifying variants predicted (by simulation) to bind roughly 20 kilocalories per mole better than their starting ancestors. This is a considerable improvement by computational or experimental standards, all at a far lower cost than standard laboratory screens.
The true promise of such tools, if borne out by experimental validation, is profound. For pharmaceutical developers, ANUBI could mean drafting higher-affinity drugs more quickly, using less physical material and fewer resources. For patients, it might, in the future, translate to more effective therapies, discovered with greater speed.
Yet, we must temper our excitement. ANUBI’s predictions, though impressively detailed, still depend on the quality of the input molecular models and simulations—a process that, like celestial navigation, can falter if the underlying maps are not accurate. Laboratory validation is still crucial, for biology always holds surprises. Still, every voyage across unexplored sequence space starts with a good map, and ANUBI promises to be a compass for the molecular explorers of tomorrow.
Sources
Buratto, D., Wang, W., Zhang, X., Zhu, Q., Meng, J., Rigden, D. J., Zhou, R., & Zonta, F. (2025). ANUBI: A Platform for Affinity Optimization of Proteins and Peptides in Drug Design. bioRxiv.

